
 | Unit Activity |
| Prev | Using NeuroScope | Next |
Unit activity is described in two files: a spike time file (listing the timestamps of the spikes) and a cluster file (assigning each spike to a cluster which corresponds to a putative unit). In addition, if you are working with a .dat file, spike waveforms can be extracted from that file. This is explained in more detail in File Formats. The spike time file and the cluster file can be loaded by selecting -> (multiple files can be opened in parallel). This creates a new cluster box in the Units Palette, where all the clusters are listed (the Units Palette is created upon loading the first cluster file).
To display the spikes fired by certain clusters, select the corresponding clusters by clicking on the colored squares in the palette. Use Shift or Ctrl for multiple selections. Clicking on a cluster file ID (the number on the left side of a unit box) will select or deselect all the clusters in the file at once. Hold the Shift key while clicking on a cluster file ID to select all clusters except 0 and 1 (artefacts and noise, respectively).
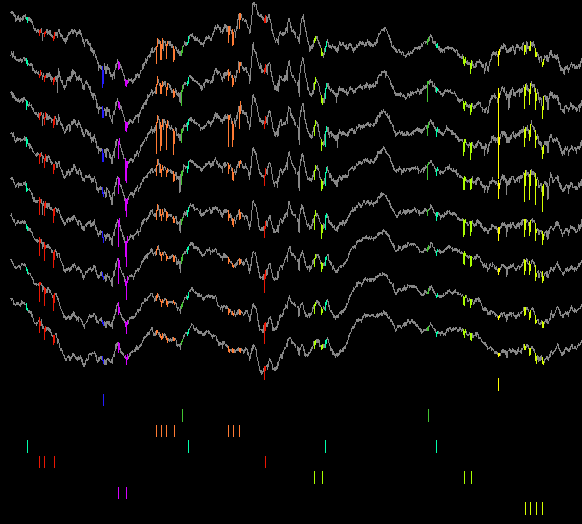
NeuroScope provides three different graphical representations for spikes. You can activate or deactivate one or more representations by selecting or deselecting the corresponding items in the menu.
Vertical Lines: each spike is represented as a vertical line spanning the entire display.
Rasters: spikes are represented as small vertical segments below the local field potential traces, grouped by unit. In the display, the total height devoted to the rasters (vs the traces) can be adjusted by selecting -> or ->.
Waveforms: spike waveforms are highlighted on their respective traces (for instance, since the .clu.5 file describes the clusters of spike group 5, selecting a cluster from this group will cause NeuroScope to draw the corresponding waveforms on the traces listed in the spike group 5). Highlighting is rendered even more visible if the traces are drawn using shades of grey instead of colors. This can be obtained by selecting ->.
The waveforms can be drawn only when working with raw data (.dat files), as the waveform information is not available in low-pass filtered data.
In multiple column mode, spikes recorded from a given set of electrodes are shown only in the column containing the corresponding traces. If however these traces are split across two or more columns, the spikes will be shown in each of these columns.
Pyramidal or granule cells typically fire at low rates. Thus, when examining their firing patterns, you may sometimes find it difficult to locate those epochs when the neurons discharge. NeuroScope provides a mechanism to easily 'browse' spikes, i.e. to automatically move the time window to the previous or next spike fired by one or more neurons. First, indicate which clusters you wish to browse by holding both Ctrl and Alt and clicking on the corresponding colored squares in the palette (only already selected clusters can be used). The squares are now replaced by triangles to reflect their activated state. Now, choose -> or click on
 to move the time window to the previous spike fired by one of the activated clusters, or choose -> or click on
to move the time window to the previous spike fired by one of the activated clusters, or choose -> or click on
 to move the time window to the next spike.
to move the time window to the next spike.
Holding Ctrl and Alt while clicking on a cluster file ID will activate or deactivate all selected clusters in the corresponding file.
Since only selected clusters can be browsed, deselecting a unit will automatically reset its activation state.
| Prev | Home | Next |
| Tools | Up | Events |